Cylindrical Fibers with step index profile
Scott Prahl
Sept 2023
Graphs and tables from Chapter 8 of Ghatak.
[1]:
# Jupyterlite support for ofiber
try:
import micropip
await micropip.install("ofiber")
except ModuleNotFoundError:
pass
import matplotlib.pyplot as plt
import numpy as np
from scipy.optimize import curve_fit
import ofiber
%config InlineBackend.figure_format='retina'
Single mode in a circular step fiber V=2
(Ghatak Fig 8.1)
Use ofiber.plot_LP_modes to make a quick plot showing the allowed modes.
[2]:
V=6.5
ell = 0
plt.subplots(1, 2, figsize=(10, 4))
plt.subplot(1, 2, 1)
aplt = ofiber.plot_LP_modes(V,ell)
plt.subplot(1, 2, 2)
ell=1
aplt = ofiber.plot_LP_modes(V,ell)
aplt.show()
for ell in range(2):
all_b = ofiber.LP_mode_values(V,ell)
for i,b in enumerate(all_b):
print("LP_%d%d, b=%.4f"%(ell,i+1,b))
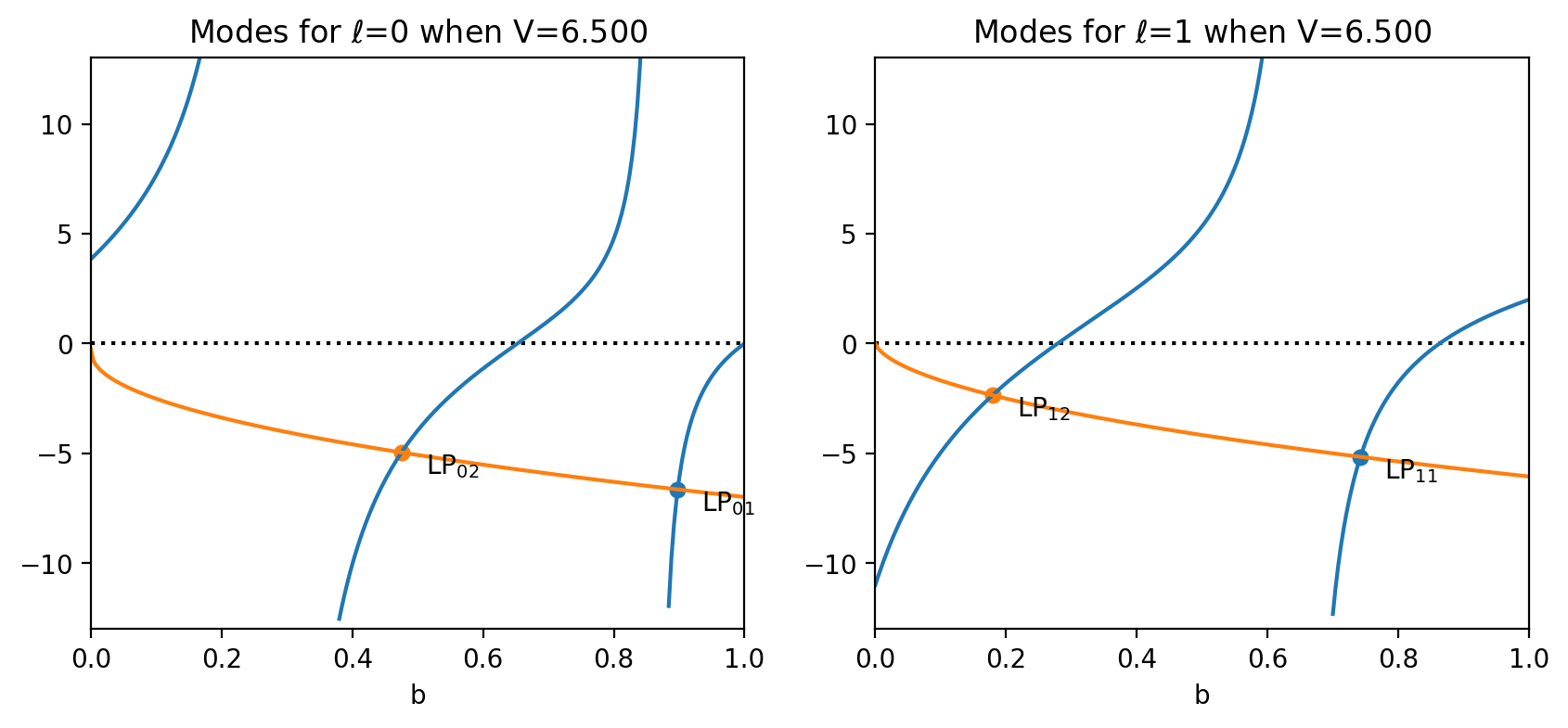
LP_01, b=0.8977
LP_02, b=0.4752
LP_11, b=0.7422
LP_12, b=0.1792
Multiple modes in a circular step fiber V=12.5
Use ofiber.plot_LP_modes to make a quick plot showing the allowed modes.
[3]:
V=12.5
ell = 0
aplt = ofiber.plot_LP_modes(V,ell)
aplt.show()
for ell in range(6):
all_b = ofiber.LP_mode_values(V,ell)
for i,b in enumerate(all_b):
print("LP_%d%d, b=%.4f"%(ell,i+1,b))
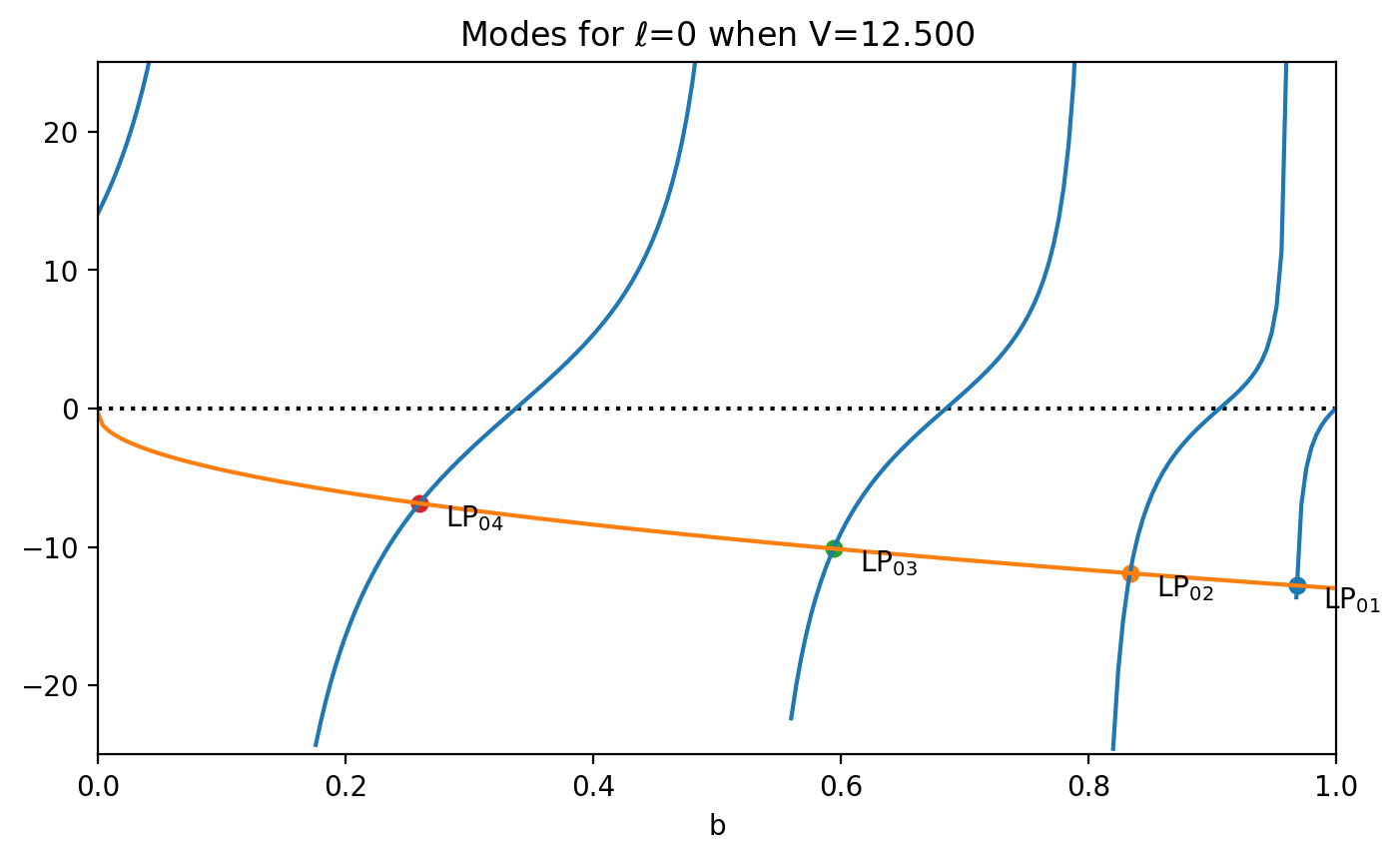
LP_01, b=0.9683
LP_02, b=0.8336
LP_03, b=0.5942
LP_04, b=0.2597
LP_11, b=0.9196
LP_12, b=0.7319
LP_13, b=0.4427
LP_14, b=0.0735
LP_21, b=0.8558
LP_22, b=0.6155
LP_23, b=0.2791
LP_31, b=0.7777
LP_32, b=0.4850
LP_33, b=0.1070
LP_41, b=0.6860
LP_42, b=0.3415
LP_51, b=0.5812
LP_52, b=0.1863
Ghatak Table 8.1
Summarizing all the important parameters for low V-parameter fibers.
[4]:
V=np.linspace(1.5,2.5,41)
ell=0
em=1
print(' V b U W wp/r_core MFD/r_core')
for v in V:
b = ofiber.LP_mode_value(v,ell,em)
W = v*np.sqrt(b)
U = v*np.sqrt(1-b)
w_p = ofiber.PetermannW(v)
w_mfd = ofiber.MFD(v)
print("%.3f %.5f %.5f %.5f %.5f %.5f"%(v,b,U,W,w_p,w_mfd))
V b U W wp/r_core MFD/r_core
1.500 0.22925 1.31689 0.71820 1.69342 3.56805
1.525 0.23955 1.32985 0.74640 1.65638 3.47716
1.550 0.24980 1.34252 0.77469 1.62172 3.39317
1.575 0.25997 1.35489 0.80305 1.58923 3.31537
1.600 0.27006 1.36698 0.83148 1.55872 3.24312
1.625 0.28007 1.37880 0.85997 1.53001 3.17585
1.650 0.28997 1.39034 0.88851 1.50295 3.11309
1.675 0.29977 1.40163 0.91709 1.47741 3.05439
1.700 0.30947 1.41267 0.94571 1.45326 2.99939
1.725 0.31905 1.42347 0.97435 1.43040 2.94774
1.750 0.32851 1.43403 1.00303 1.40872 2.89915
1.775 0.33785 1.44436 1.03172 1.38814 2.85335
1.800 0.34707 1.45448 1.06043 1.36858 2.81010
1.825 0.35616 1.46437 1.08914 1.34996 2.76920
1.850 0.36512 1.47406 1.11787 1.33223 2.73045
1.875 0.37396 1.48355 1.14660 1.31531 2.69369
1.900 0.38266 1.49285 1.17533 1.29915 2.65876
1.925 0.39123 1.50195 1.20406 1.28371 2.62552
1.950 0.39967 1.51087 1.23279 1.26893 2.59385
1.975 0.40798 1.51962 1.26150 1.25478 2.56363
2.000 0.41616 1.52818 1.29021 1.24122 2.53477
2.025 0.42421 1.53658 1.31891 1.22820 2.50718
2.050 0.43213 1.54482 1.34760 1.21571 2.48076
2.075 0.43992 1.55290 1.37627 1.20370 2.45544
2.100 0.44758 1.56082 1.40493 1.19216 2.43115
2.125 0.45512 1.56860 1.43357 1.18105 2.40783
2.150 0.46252 1.57622 1.46220 1.17035 2.38541
2.175 0.46981 1.58371 1.49080 1.16003 2.36385
2.200 0.47697 1.59106 1.51938 1.15009 2.34308
2.225 0.48401 1.59827 1.54795 1.14049 2.32308
2.250 0.49093 1.60536 1.57649 1.13122 2.30379
2.275 0.49773 1.61231 1.60501 1.12227 2.28517
2.300 0.50442 1.61915 1.63351 1.11361 2.26719
2.325 0.51099 1.62586 1.66199 1.10523 2.24981
2.350 0.51744 1.63246 1.69044 1.09713 2.23301
2.375 0.52379 1.63894 1.71887 1.08928 2.21675
2.400 0.53003 1.64531 1.74727 1.08168 2.20101
2.425 0.53616 1.65157 1.77565 1.07431 2.18576
2.450 0.54218 1.65773 1.80400 1.06716 2.17098
2.475 0.54810 1.66379 1.83233 1.06022 2.15665
2.500 0.55392 1.66974 1.86064 1.05349 2.14274
Example 8.1 Figure 8.2
[5]:
n_clad =1.45
Delta=0.0064
r_core=3.0 # microns
λ = 1.546 # microns
n_core = n_clad / np.sqrt(1 - 2*Delta)
NA = np.sqrt(n_core**2-n_clad**2)
V = r_core * (2 * np.pi / λ) * NA
plt.subplots(1, 2, figsize=(10, 4), sharey=True)
plt.subplot(1, 2, 1)
plt.plot([0,r_core,r_core,2*r_core],[n_core,n_core,n_clad,n_clad],color='blue')
for ell in range(6):
all_b = ofiber.LP_mode_values(V,ell)
for b in all_b:
bt = np.sqrt(n_clad**2+all_b*(n_core**2-n_clad**2))
plt.plot([0,r_core],[bt,bt],'--r')
plt.ylim(1.445,1.46)
plt.title("V=%.1f, λ=%.4f microns" % (V,λ))
plt.xlabel("radius (microns)")
plt.ylabel("Refractive Index")
λ = 0.4757# microns
n_core = n_clad / np.sqrt(1 - 2*Delta)
NA = np.sqrt(n_core**2-n_clad**2)
V = r_core * (2 * np.pi / λ) * NA
plt.subplot(1, 2, 2)
plt.plot([0,r_core,r_core,2*r_core],[n_core,n_core,n_clad,n_clad],color='blue')
for ell in range(6):
all_b = ofiber.LP_mode_values(V,ell)
for b in all_b:
bt = np.sqrt(n_clad**2+all_b*(n_core**2-n_clad**2))
plt.plot([0,r_core],[bt,bt],'--r')
plt.ylim(1.445,1.46)
plt.title("V=%.1f, λ=%.4f microns" % (V,λ))
plt.xlabel("radius (microns)")
#plt.ylabel("Refractive Index")
plt.show()
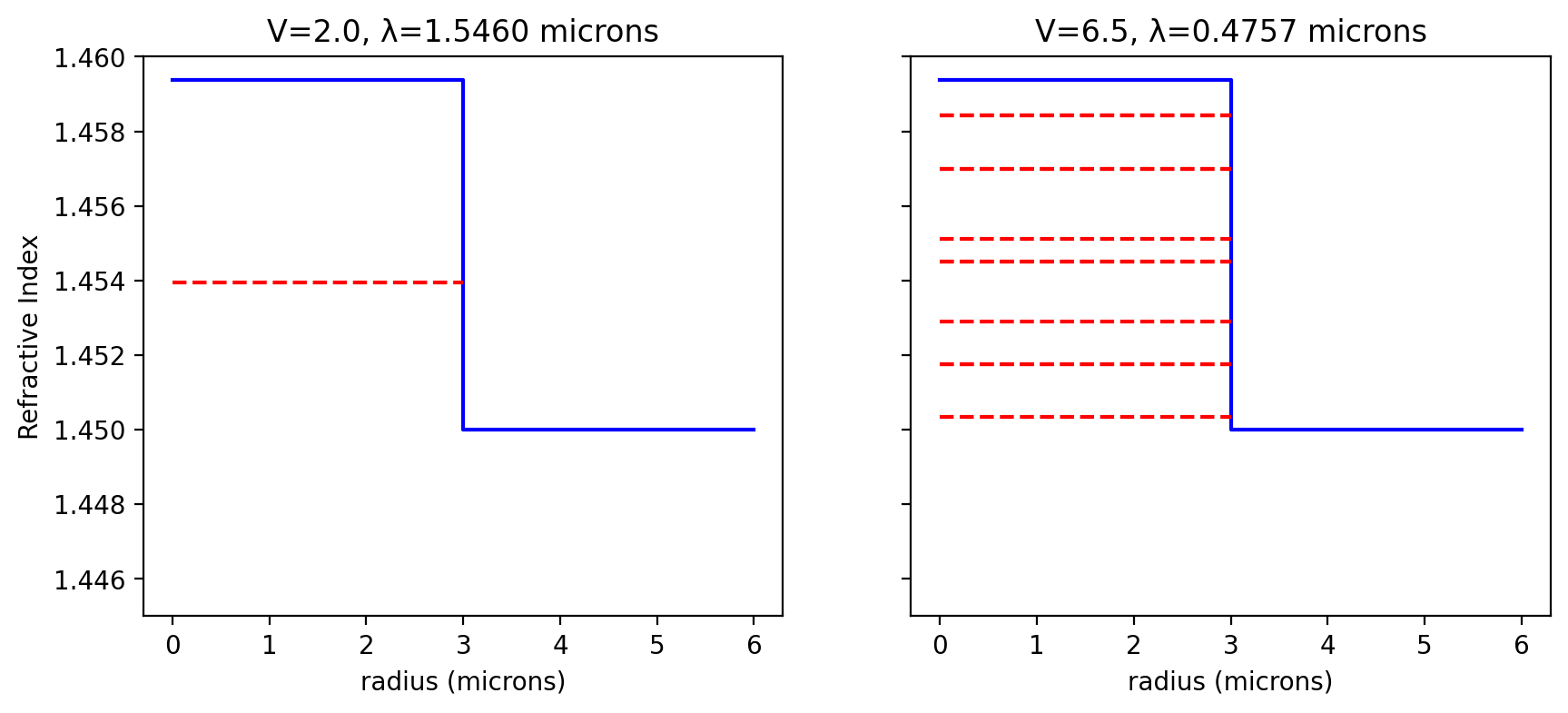
Figure 8.3
[6]:
n_clad =1.45
Delta=0.010
r_core=2.0 # microns
λ = 1.288 # microns
n_core = n_clad / np.sqrt(1 - 2*Delta)
NA = np.sqrt(n_core**2-n_clad**2)
V = r_core * (2 * np.pi / λ) * NA
plt.subplots(1, 2, figsize=(10, 4), sharey=True)
plt.subplot(1, 2, 1)
plt.plot([0,r_core,r_core,2*r_core],[n_core,n_core,n_clad,n_clad],color='blue')
for ell in range(6):
all_b = ofiber.LP_mode_values(V,ell)
for b in all_b:
bt = np.sqrt(n_clad**2+all_b*(n_core**2-n_clad**2))
plt.plot([0,r_core],[bt,bt],'--r')
plt.ylim(1.445,1.47)
plt.title("V=%.1f, λ=%.4f microns" % (V,λ))
plt.xlabel("radius (microns)")
plt.ylabel("Refractive Index")
λ = 0.3963# microns
n_core = n_clad / np.sqrt(1 - 2*Delta)
NA = np.sqrt(n_core**2-n_clad**2)
V = r_core * (2 * np.pi / λ) * NA
plt.subplot(1, 2, 2)
plt.plot([0,r_core,r_core,2*r_core],[n_core,n_core,n_clad,n_clad],color='blue')
for ell in range(6):
all_b = ofiber.LP_mode_values(V,ell)
for b in all_b:
bt = np.sqrt(n_clad**2+all_b*(n_core**2-n_clad**2))
plt.plot([0,r_core],[bt,bt],'--r')
plt.ylim(1.445,1.47)
plt.title("V=%.1f, λ=%.4f microns" % (V,λ))
plt.xlabel("radius (microns)")
#plt.ylabel("Refractive Index")
plt.show()
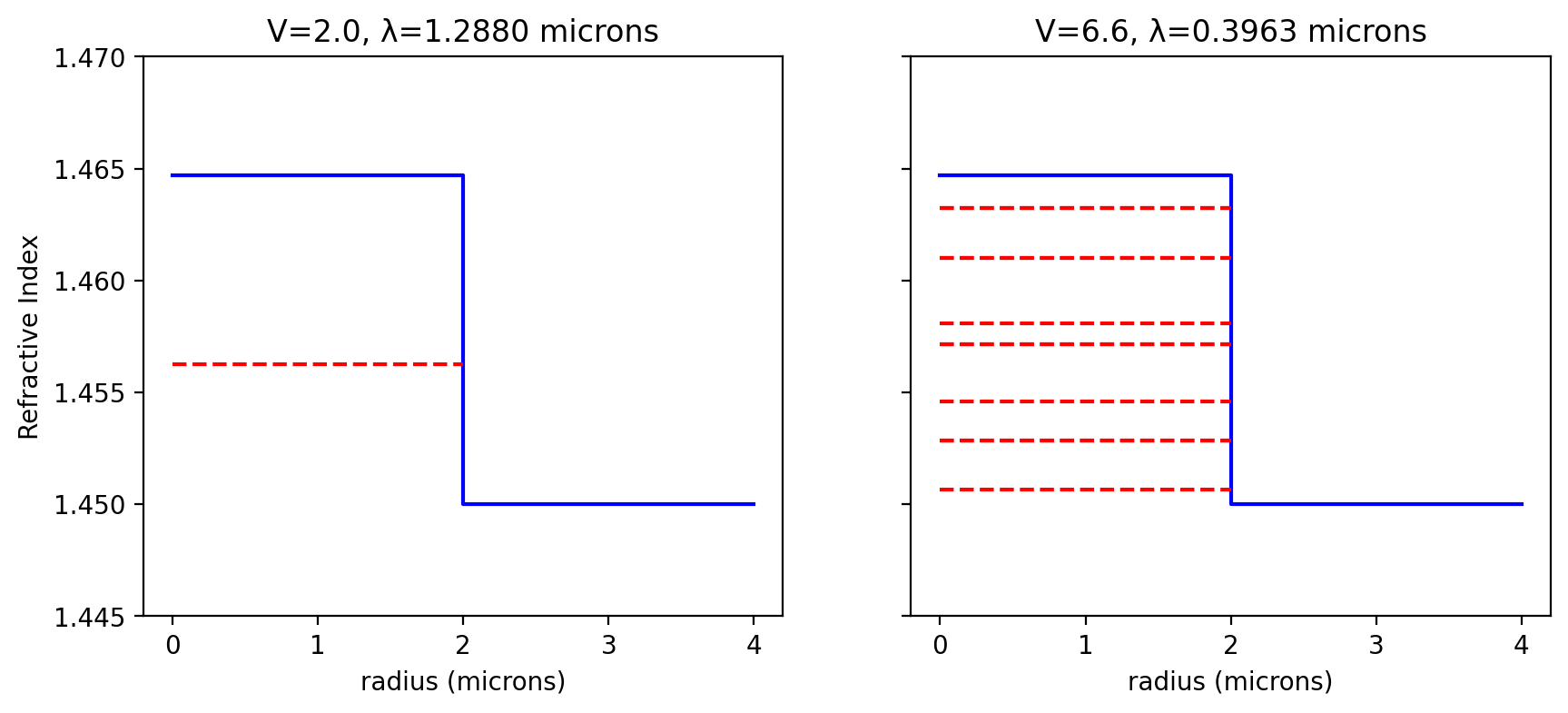
Ghatak Figure 8.4
[7]:
V=6.5
plt.subplots(3, 2, figsize=(15, 15),squeeze=True)
for ell in range(6):
aplt.subplot(3,2,ell+1)
aplt = ofiber.plot_LP_modes(V,ell)
aplt.show()
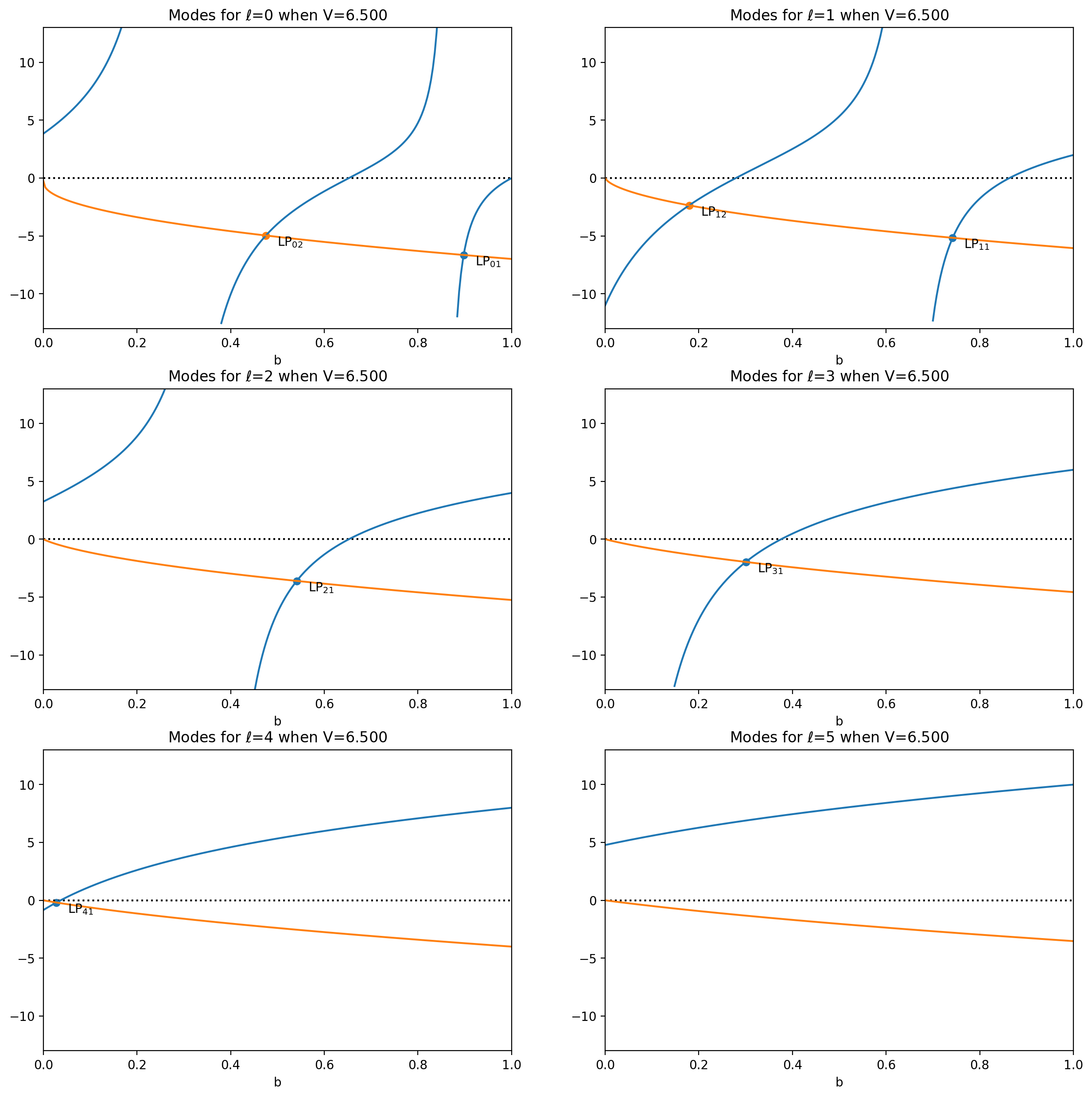
Variation of normalized propagation constant with V
Ghatak Figure 8.5
[8]:
V = np.linspace(0.01,10,50)
b = np.empty_like(V)
for ell in range(3):
for em in range(1,4):
for i in range(len(V)):
b[i] = ofiber.LP_mode_value(V[i],ell,em)
plt.plot(V,b)
plt.annotate(r' LP$_{%d%d}$'%(ell,em), xy=(10,b[-1]),va='center')
plt.xlim(0,12)
plt.xlabel("V-parameter")
plt.ylabel('Propagation Constant b')
plt.title('Modes in a step-index Fiber')
plt.show()
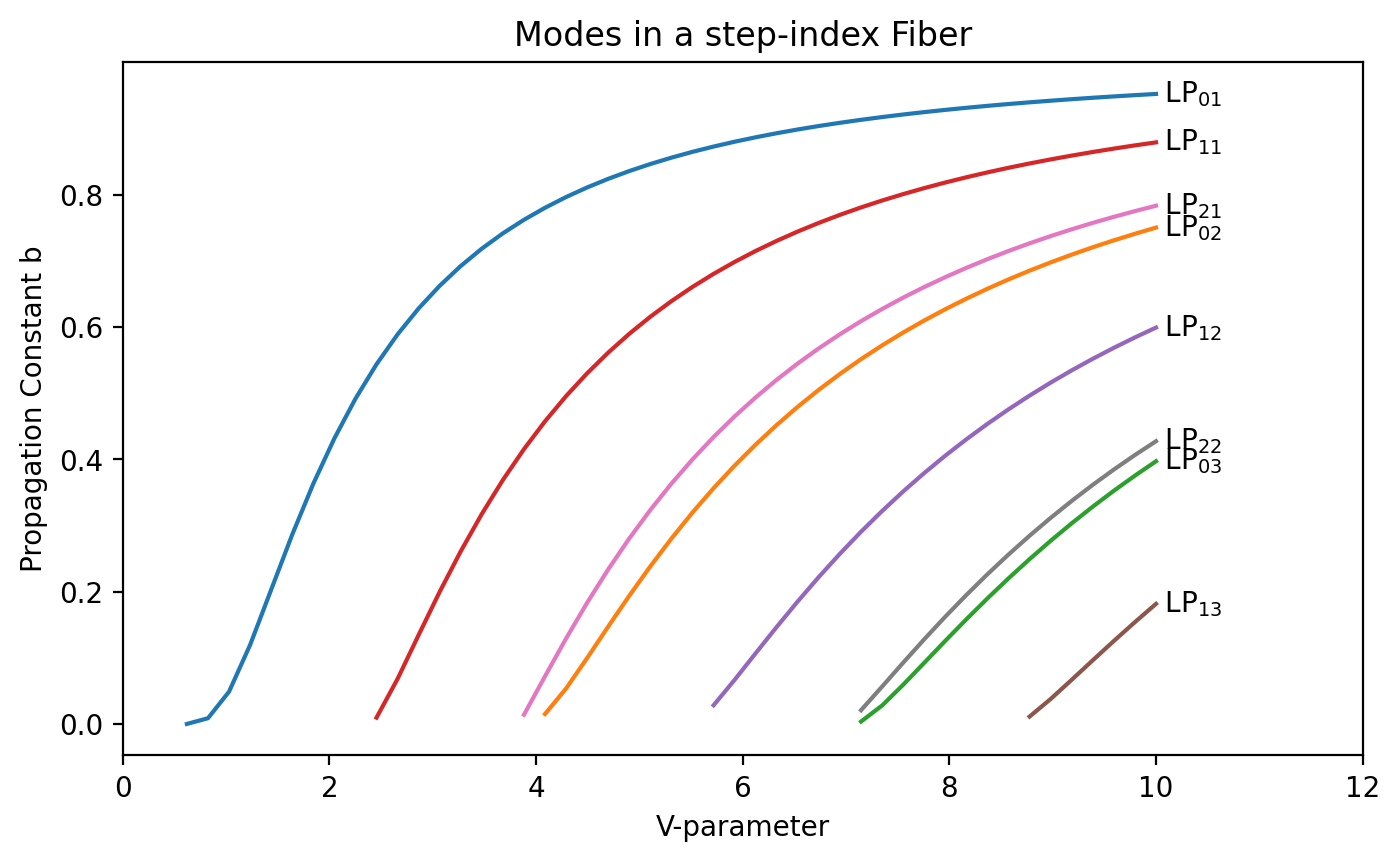
Radial Intensity distributions
Ghatak Figure 8.6
[9]:
plt.subplots(figsize=(10,6))
r_over_a = np.linspace(-1.2,1.2,100)
V=8
for ell in range(3):
for em in range(2,4):
b = ofiber.LP_mode_value(V,ell,em)
if b is None :
continue
irradiance = ofiber.LP_radial_irradiance(V,b,ell,r_over_a)
i = np.argmax(irradiance)
plt.plot(r_over_a,irradiance)
plt.annotate(r' LP$_{%d%d}$'%(ell,em), xy=(r_over_a[i],irradiance[i]),va='bottom',ha='center')
plt.xlabel('r/r_core')
plt.ylabel('Normalized Irradiance')
plt.plot([-1,-1],[0,10],':k')
plt.plot([1,1],[0,10],':k')
plt.title('Step-index fiber, V=%.2f'%V)
plt.ylim(0,10)
plt.xlim(-1.2,1.2)
plt.show()
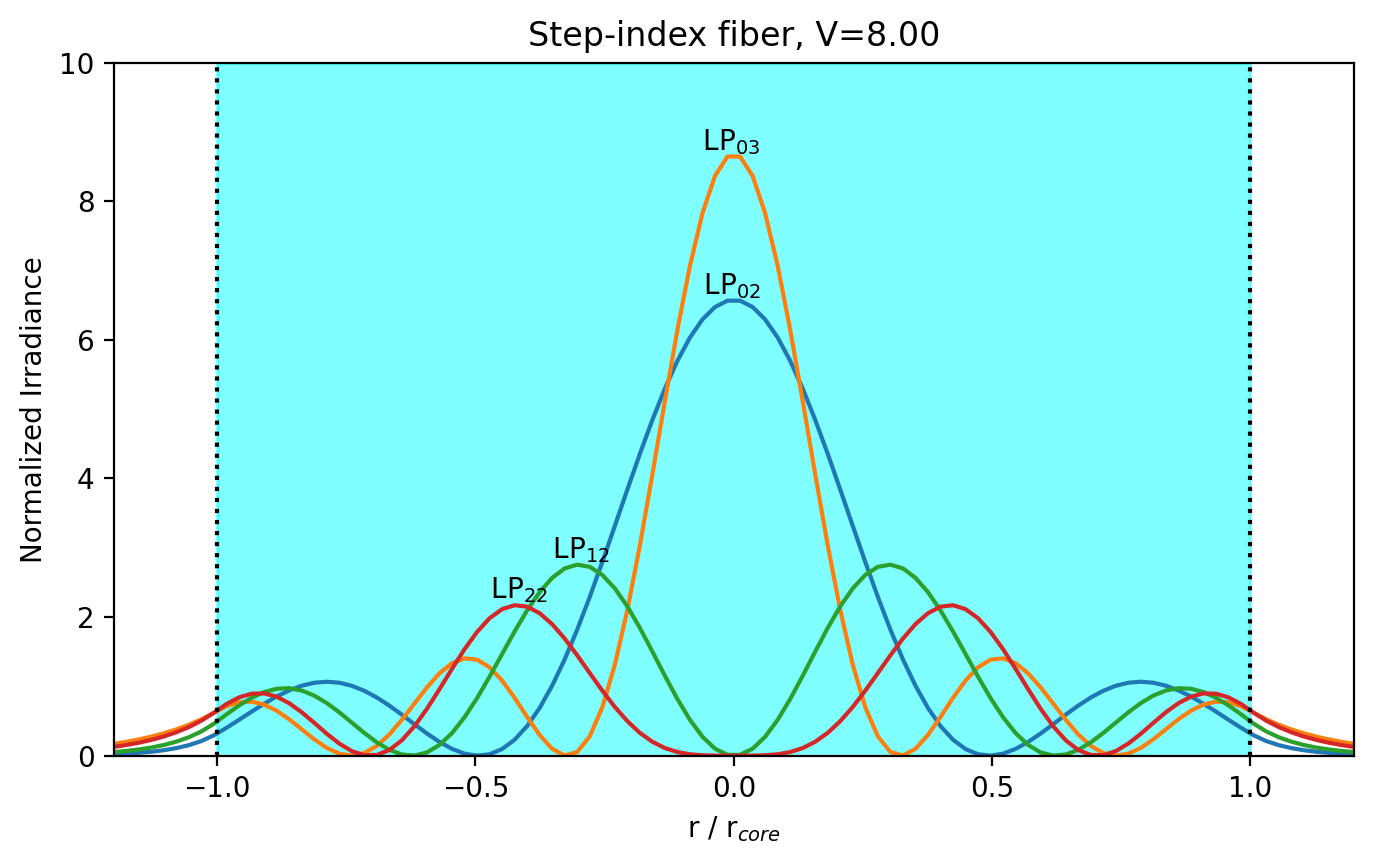
[10]:
V=8
r_over_a = np.linspace(0, 1.5, 50)
phi = np.linspace(0, 2*np.pi, 40)
clevs = np.linspace(-4, 4, 9)
fig,ax=plt.subplots(3,3,figsize=(8,8))
graph = 0
for ell in [0,1,2]:
for em in [1,2,3]:
graph = graph+1
b = ofiber.LP_mode_value(V,ell,em)
if b is None :
ax=plt.subplot(3,3,graph,polar=True)
plt.axis('off')
plt.annotate('No such mode', xy=(0,0), ha='center')
continue
r_field = ofiber.LP_radial_field(V,b,ell,r_over_a)
phi_field = np.cos(ell*phi)
R, PHI = np.meshgrid(r_over_a, phi)
R_FIELD, PHI_FIELD = np.meshgrid(phi_field,r_field)
Z = R_FIELD*PHI_FIELD
ax=plt.subplot(3,3,graph,polar=True)
cax=ax.contourf(phi,r_over_a,Z,levels=clevs)
ax.set_xticklabels([])
ax.set_yticklabels([])
fig.colorbar(cax)
plt.title(r"LP$_{%d%d}$ Field"%(ell,em))
plt.show()
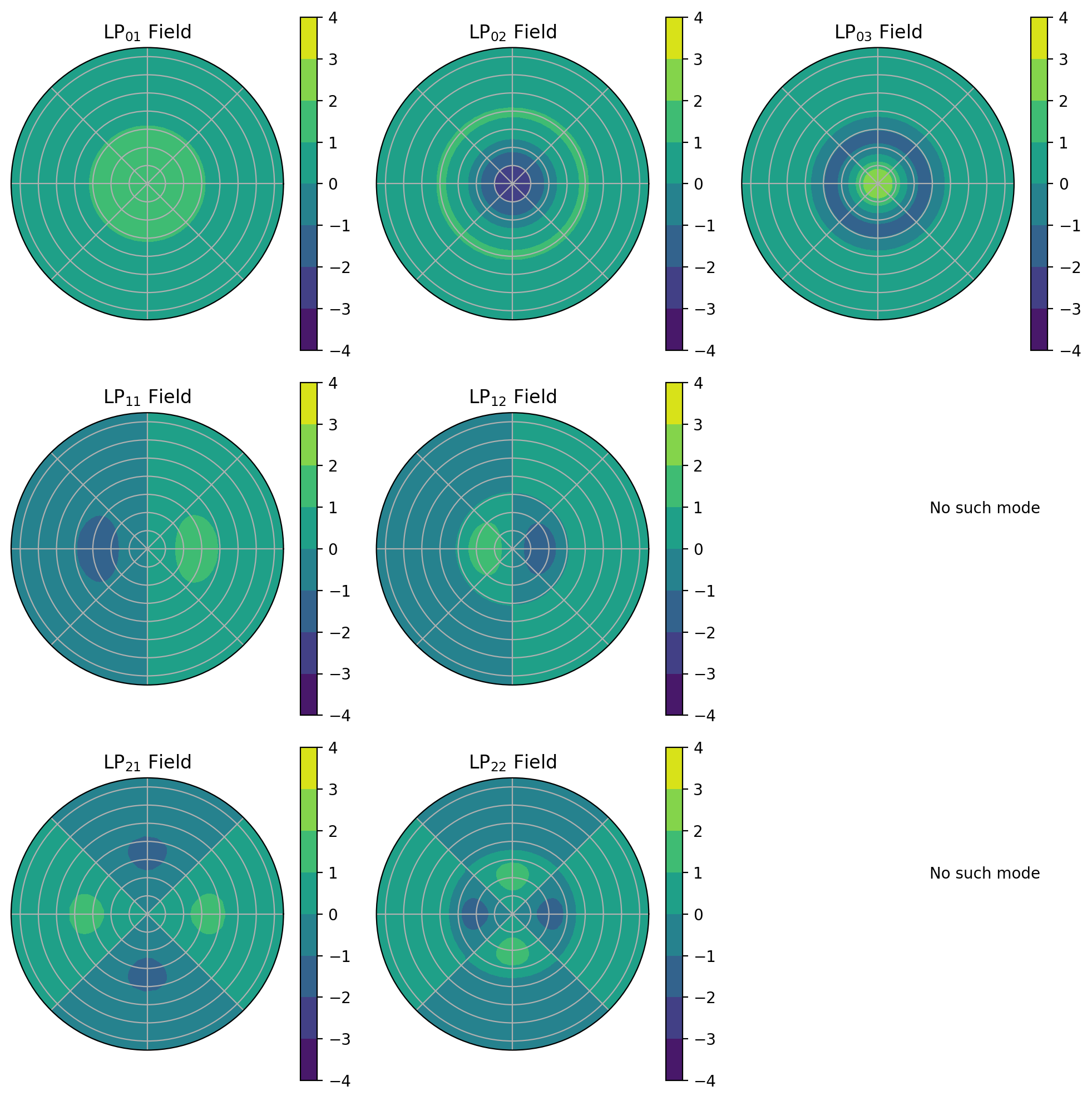
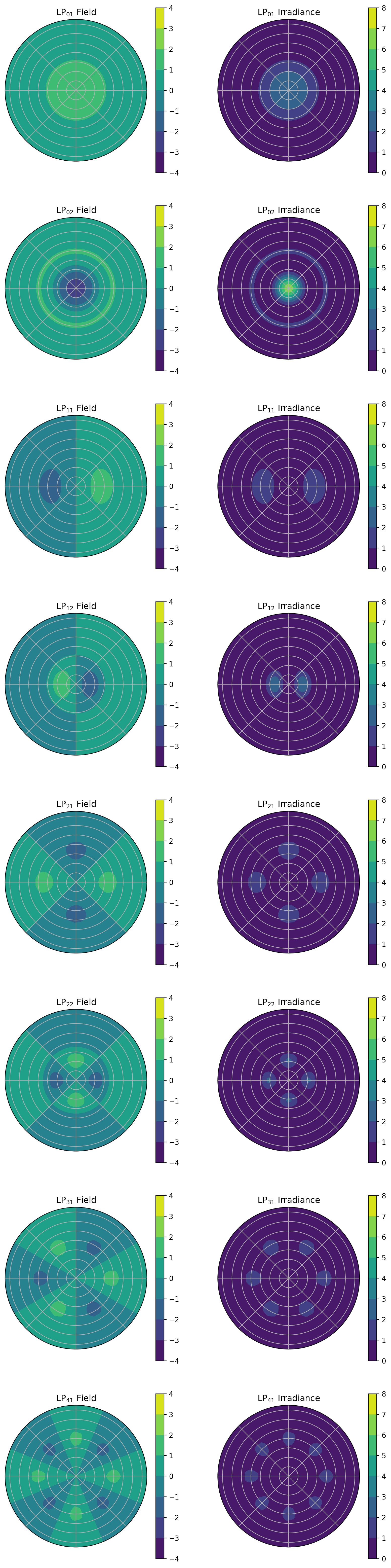
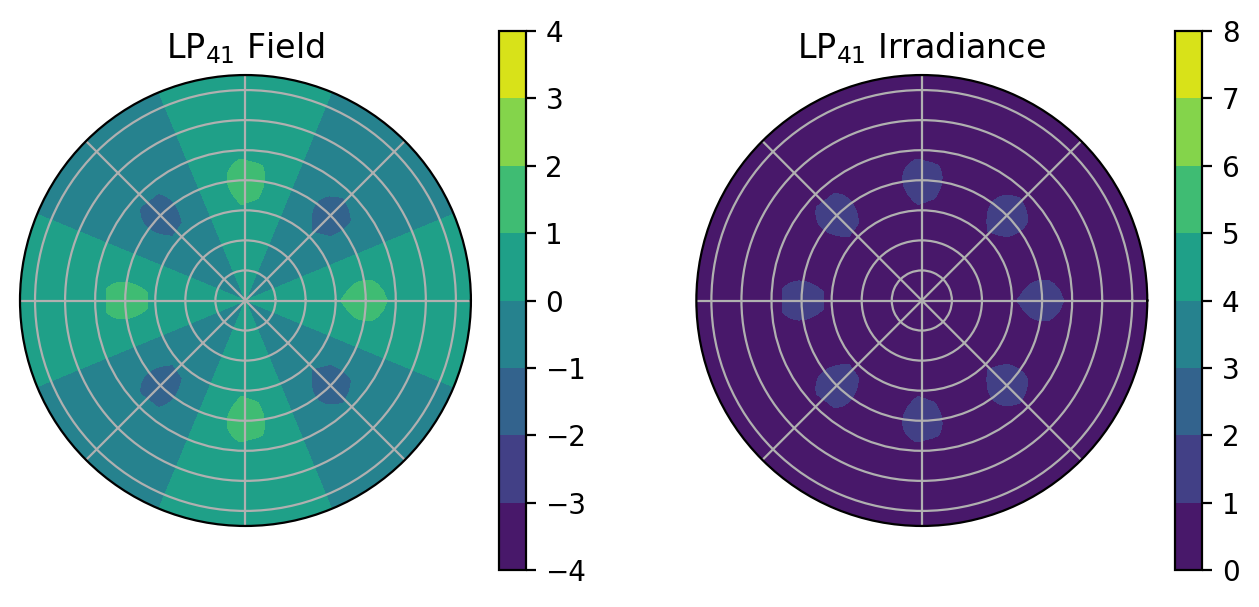
[11]:
V=8
r_over_a = np.linspace(0, 1.5, 50)
phi = np.linspace(0, 2*np.pi, 40)
R, PHI = np.meshgrid(r_over_a, phi)
clevs = np.linspace(-4, 4, 9)
dlevs = np.linspace(0, 8, 9)
fig,ax=plt.subplots(figsize=(10,40))
graph = 0
rows = 8
cols = 2
for ell in range(5):
phi_field = np.cos(ell*phi)
for em in [1,2]:
b = ofiber.LP_mode_value(V,ell,em)
if b is None:
continue
r_field = ofiber.LP_radial_field(V,b,ell,r_over_a)
R_FIELD, PHI_FIELD = np.meshgrid(phi_field,r_field)
Z = R_FIELD*PHI_FIELD
graph += 1
ax=plt.subplot(rows,cols,graph,polar=True)
cax=ax.contourf(phi,r_over_a,Z,levels=clevs)
ax.set_xticklabels([])
ax.set_yticklabels([])
fig.colorbar(cax)
plt.title(r"LP$_{%d%d}$ Field"%(ell,em))
graph += 1
Z = Z*Z
ax=plt.subplot(rows,cols,graph,polar=True)
cax=ax.contourf(phi,r_over_a,Z,levels=dlevs)
ax.set_xticklabels([])
ax.set_yticklabels([])
fig.colorbar(cax)
plt.title(r"LP$_{%d%d}$ Irradiance"%(ell,em))
plt.show()
[12]:
def gauss(x, A, w):
return A*np.exp(-x**2/w**2)
r_over_a = np.linspace(-1.5, 1.5, 50)
phi = 0
graph = 1
rows = 4
cols = 1
fig,ax=plt.subplots(figsize=(6,18))
ell = 0
em = 1
for V in [2.4,3,3.4,4]:
b = ofiber.LP_mode_value(V,ell,em)
field = ofiber.LP_radial_field(V,b,ell,r_over_a)
plt.subplot(rows,cols,graph)
plt.plot(r_over_a,field,label=r'LP$_{01}$')
plt.annotate("V=%.2f"%V, xy=(-1.5,max(field)))
popt, pcov = curve_fit(gauss, r_over_a, field)
fit = gauss(r_over_a,popt[0],popt[1])
plt.plot(r_over_a,fit,':r',label='fit')
plt.ylabel('Normalized Field')
plt.xlabel('r/r_core')
plt.legend()
graph += 1
plt.show()
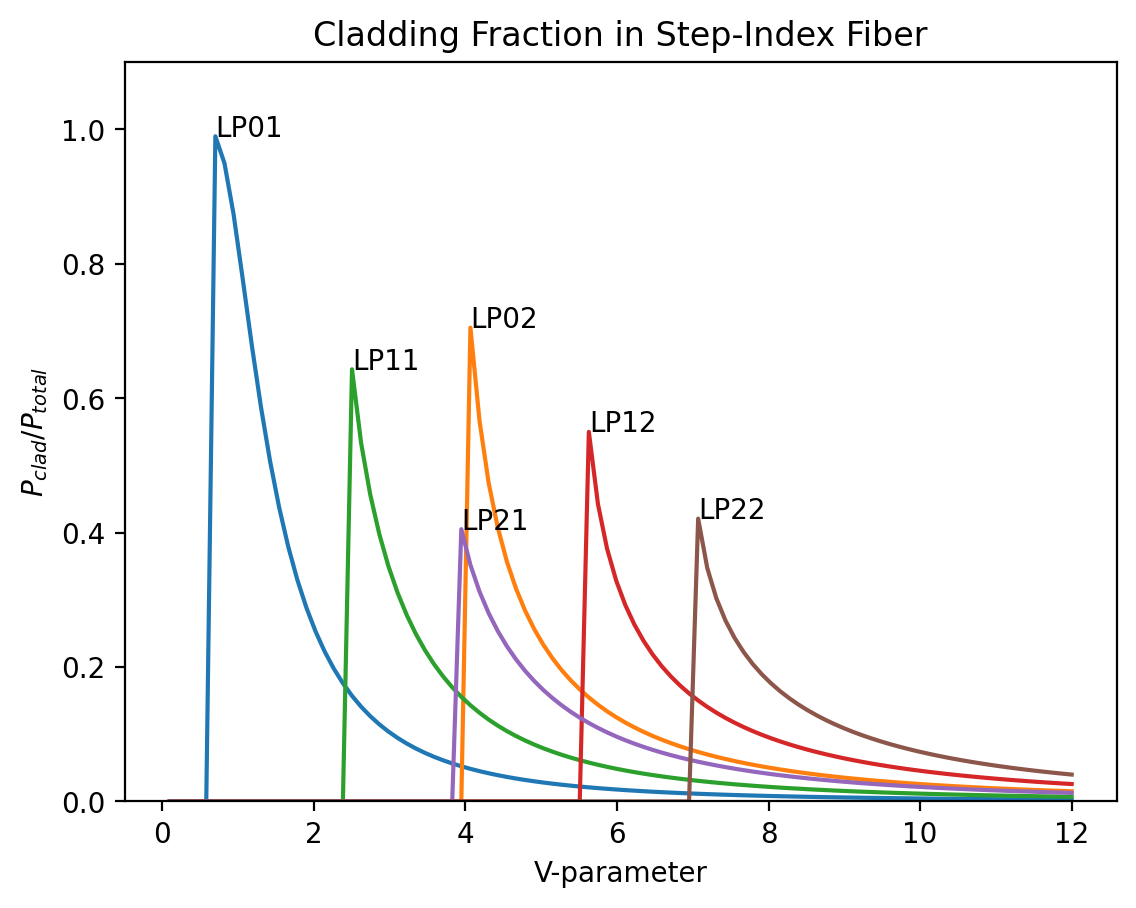
Variation of the fractional power contained in the cladding with V
Ghatak Figure 8.11
[13]:
V=np.linspace(0.1,12,100)
ell = 0
for ell in [0,1,2]:
for em in [1,2]:
ratio = np.zeros_like(V)
for i in range(len(V)):
b = ofiber.LP_mode_value(V[i],ell,em)
if b is None:
continue
clad = ofiber.LP_clad_irradiance(V[i],b,ell)
total = ofiber.LP_total_irradiance(V[i],b,ell)
ratio[i] = clad/total
j = np.nanargmax(ratio)
plt.annotate("LP%d%d"%(ell,em),xy=(V[j],ratio[j]))
plt.plot(V,ratio)
plt.xlabel('V-parameter')
plt.ylabel(r'$P_{clad}/P_{total}$')
plt.ylim(0,1.1)
plt.title('Cladding Fraction in Step-Index Fiber')
plt.show()
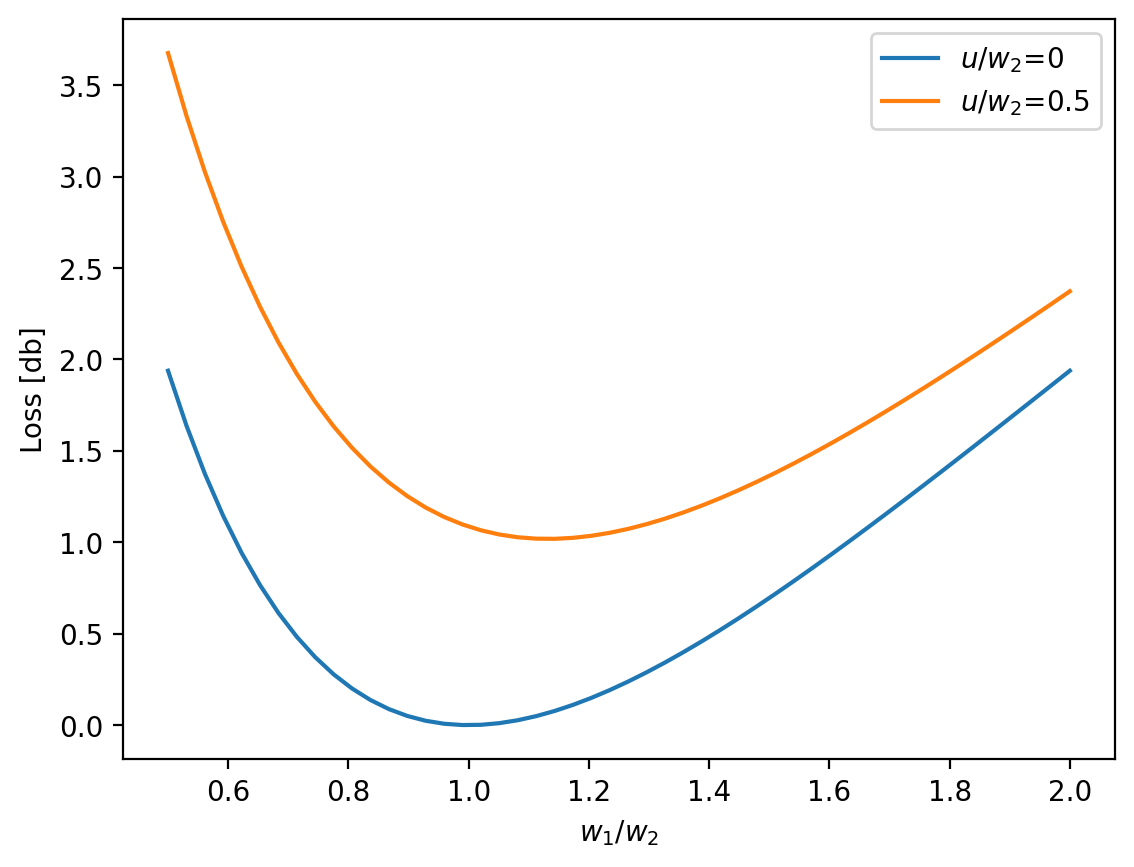
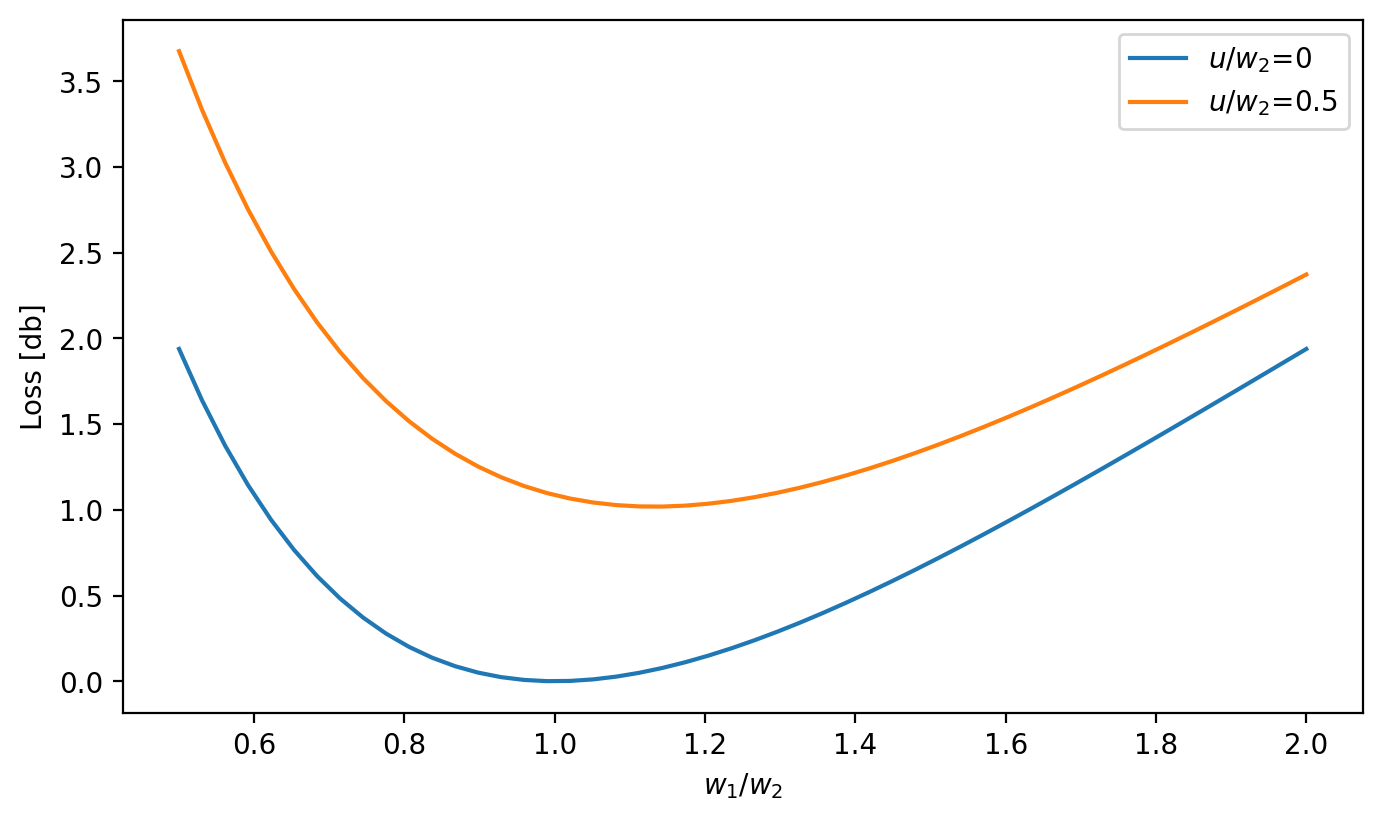
Variation of loss at a perfectly aligned joint between two single-mode fibers
Ghatak Figure 8.14
[14]:
w1 = np.linspace(0.5,2,50)
w2 = np.ones_like(w1)
loss_db = ofiber.transverse_misalignment_loss_db(w1,w2,0)
plt.plot(w1,loss_db,label=r'$u/w_2$=0')
loss_db = ofiber.transverse_misalignment_loss_db(w1,w2,0.5)
plt.plot(w1,loss_db,label=r'$u/w_2$=0.5')
plt.xlabel(r'$w_1/w_2$')
plt.ylabel('Loss [db]')
plt.legend()
plt.show()
[15]:
u = np.linspace(0,1,50)
loss_db = ofiber.transverse_misalignment_loss_db(1,1,u)
plt.plot(u,loss_db)
plt.xlabel(r'$u/w$')
plt.ylabel('Loss [db]')
plt.title(r'$w_1=w_2=w$')
plt.show()
[ ]: